Supervised by: Alexandra Knighton, BA (Oxon). Alex is a 5th year medical student at the University of Oxford who recently completed her undergraduate degree in medical sciences. Alex particularly enjoyed studying Neuroscience, with an especial interest in psychiatry, degenerative disease and circadian rhythms. She has also conducted a research project looking at ethics in global health.
Abstract
Parkinson’s disease (PD) is an age-related, progressive neurodegenerative disorder that involves the degeneration of dopaminergic neurons in the substantia nigra compacta region (SNc). This paper reviews the role of different mutations on the development of PD and discusses the role of RNAi, CRISPR and single-cell RNA sequencing in the detection and treatment of PD. LRRK2 and PINK1 mutations will be discussed. Mutations in the LRRK2 gene are associated with neuronal toxicity, and genetically interact with the TOR pathway, which drives apoptosis in dopaminergic neurons. PINK1 (PTEN induced kinase 1) mutations are commonly associated with familial PD. This gene is involved with mitochondria. RNAi interference is a currently studied treatment for PD consisting of small pieces of RNA that prevent protein translation by degrading a targeted mRNA. RNAi studies have been made on the gene that codes for parkin which showed benefits in targeting mitochondrial accumulation of the disease. Other studies have been done on SNCA using an adeno-associated virus (AAV) in rats which showed a large reduction of SNCAmRNA expression in the substantia nigra. Another studied treatment is CRISPR/Cas9 a naturally occurring system that can be manipulated to modify specific sequences of DNA, its accuracy in modifying specific DNA sequences could have potential implications for hereditary PD prevention, and PD treatments.
Introduction
Parkinson’s disease (PD) is an age-related, progressive neurodegenerative disorder that involves the degeneration of dopaminergic neurons in the substantia nigra pars compacta region (SNc) (Correddu and Leung, 2019; Pszdyshev et al.). Currently, approximately 1-4% of individuals over the age of 65 are affected by PD worldwide (Healy et al., 2008; 2017; Kumar, 2010). Clinically, PD is characterized by bradykinesia and a combination of motor symptoms such as a 5Hz tremor, rigidity, postural instability, and shuffling gait, as well as non-motor symptoms such as salivation, constipation, depression and dementia, amongst others (Maged M. et al., 2011). Whilst all cases of PD involve the degeneration of dopaminergic neurons and bradykinesia, the combination and severity of symptoms as well as the disease progression are individual to each patient. Pathologically, PD is associated with alpha-synuclein, a misfolded protein, which aggregates to form Lewy bodies (Maged M. et al., 2011). It is important to note however, that the exact nature of Lewy bodies’ influence on Parkinson’s is unknown as of now. Moreover, there is a genetic element to PD, where some cases of PD are associated with certain mutations, which may in some instances also be influenced by the environment. Notably, certain mutations are commonly associated with PD. These include mutations in the genes: LRRK2, PARK7, PINK1, PRKN, DJ1, GBA, SNCA.
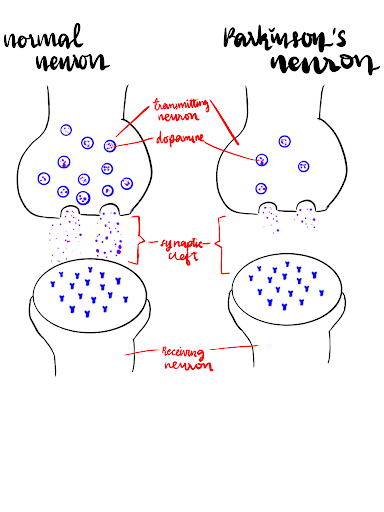
Figure 1: A diagram showing the transmission of dopamine in a normal neuron (left) and a PD neuron (right).
Currently, there is no cure for PD, however, symptomatic treatments can be used to control symptoms (Deng et al., 2008). Treatments can include medications such as Levodopa and Carbidopa, and occupational therapy and physical therapy may be used to help improve movement and speech abilities (Salat and Tolosa, 2013). It is worth noting however that these medications, while providing symptomatic relief, are often associated with significant and sometimes intolerable side-effects. Levodopa is converted to dopamine in the brain while carbidopa ensures that the levodopa does not break down before it reaches the brain. There is also neuronal apoptosis leading to fewer dopaminergic neurons. The structure of a normal and pathological neuron in a person without and with Parkinson’s respectively:
Figure 1 (Left) illustrates that in a normal neuron there is plenty of dopamine in the transmitting neuron all of which is released into the synaptic cleft and then reaches the receiving neuron.
Figure 1 (Right) shows that there is very little dopamine in a Parkinson’s neuron. Death of dopaminergic neurons may occur due to enzymatic and non-enzymatic oxidation of dopamine which generates a reactive oxygen species, inducing apoptotic cell death in dopamine neurons (Naoi et al., 1999).
The different mutations of Parkinson’s: How do these cause the individual symptoms of PD?
Protein homeostasis is achieved through a careful balance of the synthesis, folding and degradation of proteins. The regulation of protein synthesis is key to an organism’s development and homeostasis in response to a rapidly changing environment. The deregulation of protein synthesis has been associated with the development of neurodegenerative diseases such as PD. Current research supports that the misfolding and aggregation of proteins play a central role in disease development. Hence, a malfunctioning post-translational modification mechanism can result in detrimental causes such as the development of PD.
Mutations have been shown to play a central role in the pathogenesis of PD. Different mutations hypothesized to be linked to PD were identified through genome-wide association studies, including LRRK2, PARK7 (DJ-1), PINK1, PRKN, GBA, SNCA, SYCN. These mutations are linked to PD in some cases, however it is important to note that the occurrence of a mutation does not necessarily result in the development of PD in all patients as there are other factors such as the environment that influence PD. Throughout this research paper, the effects of the LRRK2 and PINK1, and their effect on PD will be reviewed individually.
SNCA: SNCA makes the protein alpha-synuclein. In the neurons of individuals with Parkinson’s disease, this protein aggregates to form Lewy bodies (Konno et al.2016). This has been explained earlier, mutations in the SNCA gene occur in early-onset Parkinson’s disease. A number of mutations in this protein, including A30P and A53T, are associated with familial forms of the disease. This aggregation has seemed to be induced by abnormal protein-protein or protein-membrane interactions by malfunction of molecular protectors and by the effect of modifications occurring after the genetic translation (Perni et al.).
PARK2: The PARK2 gene makes the protein parkin, which normally helps cells break down and recycle proteins. Mutations of the PARK2 gene are associated with mitochondrial dysfunction, leading to neuronal death in Parkinson’s (Hatano, Taku & Ueno, Shinichi & Okuzumi, Ayami & Mori, Akio & Oji, Yutaka & Hattori, Nobutaka et al., 2022).
PARK7: Mutations in this gene cause a rare form of early-onset Parkinson’s disease. The PARK7 gene makes the protein DJ-1, which protects against mitochondrial stress and is essential for nerve cell health (Heremans et al., 2022). Mutations in the gene lead to a reduction or loss of the production of DJ-1 and are associated with early onset Parkinson’s. However, how this change triggers the death of nerve cells that produce the signalling molecule dopamine — the underlying cause of Parkinson’s disease — is still unclear. (PhD Bryson et al. 2020)
PINK1: The protein made by PINK1 is a protein kinase that protects mitochondria from stress. PINK1 mutations occur in early-onset Parkinson’s disease (Pickrell and Youle et al., 2015).
LRRK2: The protein made by LRRK2 is also a protein kinase. Mutations in the LRRK2 gene have been linked to late-onset Parkinson’s disease (Usmani et al., 2021), both in the sporadic and familial aspect (Deng et al., 2007; Martin et al., 2014).
Among inherited cases of Parkinson’s, the inheritance patterns differ depending on the genes involved. If the LRRK2 or SNCA genes are involved, Parkinson’s is likely inherited from one parent, in an autosomal dominant pattern; if the PARK2, PARK7 or PINK1 gene is involved, it’s typically in an autosomal recessive.
Mutations in the gene for a protein called Parkin are known to cause the death of dopamine neurons; less commonly, defects in another protein, PINK1, can have the same effect.
In their 2011 study, Dawson and his collaborators used mice and human brain tissue to find that Parkin adds a chemical tag known as ubiquitin to PARIS that signals other proteins to break it down.
To find out whether PINK1 and PARIS have a similar relationship, the researchers ran biochemical tests on purified proteins which revealed that PINK1 and PARIS interact. Dawson says the results revealed that while PINK1’s normal role is to add a chemical tag known as a phosphate group to a certain spot on the PARIS protein, defective forms of PINK1 linked to Parkinson’s disease cannot add that tag.
Dawson said that the phosphate addition kicks off a chain of events that ultimately leads to the dismantling of the PARIS protein, a cause-and-effect relationship his team verified by reducing the amount of PINK1 in lab-grown human cells, which led to a threefold increase in the amount of PARIS. Similarly, reducing the amount of PINK1 made in living mice by more than 80 percent led to a doubling in the amount of PARIS present.
Discussed below are the two most frequent mutations seen in familial Parkinson’s disease:
The LRRK2 mutation
LRRK2 (leucine-rich repeat kinase 2) is a large multidomain protein with enzymatic RAS-link GTPase and kinase domains (Danilo, 2019; Kumar et al., 2010; Martin, 2016). LRRK2 has been hypothesized to play a role in several fundamental cellular processes, including: autophagy, endocytosis and mitochondrial and cytoskeletal function (Lopez et al., 2020). Mutations in the LRRK2 gene are the most common PD mutation and are commonly associated with an age-related familial PD as well as in sporadic PD, in which the mutant LRRK2 acts autosomally dominant (Deng et al., 2007; Martin et al., 2014). Notably, familial PD is commonly caused by mutations in the catalytic Ras in Complex domain (Roc-COR), which belongs to the GTPase superfamily that catalyse the hydrolysis of guanosine triphosphate (GTP) to guanosine diphosphate (GDP) and inorganic phosphate (Bosgraaf and Van Hasstert, 2003; Martin et al., 2014; Lopez et al., 2020). Clinically, LRRK2 mutations present themselves with neurodegeneration, and a clinical presentation similar to that of late-onset PD (Martin, 2016). Lewy body pathology is not always present: Lewy body pathology is mainly present in G2019S mutation, whilst other LRRK2 mutations (R1441C, Y1699C, and I2020T) frequently do not exhibit Lewy body pathology (Martin et al., 2014; Martin, 2016). This lack of Lewy body pathology in certain mutations increases the difficulty surrounding the diagnosing and treatment of PD as carbidopa-levodopa, which usually targets Lewy body pathology is ineffective in such patients.
The G2019S mutation in LRRK2’s kinase domain is of notable prevalence: 4% of familial cases and 1% of sporadic cases PD display LRRK2 mutations (Healy et al., 2008; Martin et al., 2014). In the G2019S mutation in LRRK2 there is an enhanced kinase activity that results in an increased phosphorylation, associated with neuronal toxicity in both in vitro and in vivo models (Danilo, 2019; Martin, 2016). Notably, G2019S and I2020T mutations in the LRRK2 gene were found to enhance the phosphorylation of the ribosomal protein s15 and this stimulated both cap-dependent and cap-independent mRNA translation in an s15-dependent manner (Martin, 2016). This was identified as a mediator of mutant LRRK2 toxicity in primary neuron and Drosophila models (Martin, 2016). However, the manner in which s15 phosphorylation affects mRNA translation is currently unresolved (Martin, 2016). A similar effect was seen in the I2020T mutation, which was also associated with an increase in kinase activity (Martin, 2016). On the contrary, it is less clear how the Roc-COR mutations affect LRRK2 kinase activity (Martin, 2016).
In studies on Drosophila expressing kinase-enhancing dLRRK mutations, findings suggested that LRRK2 genetically interacts with the target of rapamycin (TOR) pathway (Martin, 2016; Tain et al., 2009). The loss of normal LRRK2 leads to downregulated TOR signalling caused by the overexpression of a constitutively active form of the eIF4E-binding protein 1 (4E-BP1) or overexpression of tuberous sclerosis complex 1 (TSC1) and tuberous sclerosis complex 2 (TSC2) (Martin, 2016). The stimulatory effects of TOR signalling on 5’ cap-dependent translation are mediated via the phosphorylation of 4E-BP1 which disrupts its eIF4E binding activities (Danilo, 2019; Martin et al., 2014; Martin, 2016; Tain et al., 2009). This promotes cap complex formation through the binding of eIF4G to free eIF4E (Danilo, 2019; Martin, 2016). The downregulated TOR signalling enhances tau-induced neurodegeneration in a cell-cycle dependent manner, which drives apoptosis in dopaminergic neurons and results in the neurodegenerative aspect of PD (Khurana et al., 2006).
Mutations in the LRRK2 genes in both humans and Drosophila were associated with the phosphorylation of 4E-BP1, which in turn promoted PD-related phenotypes via eIF4E-dependent translation initiation (Danilo, 2019; Martin, 2016). Contrastingly, in G2019S knock-in mice, an increased phosphorylation of 4E-BP1 was not found, but was reported in G2019S knock-out mice (Martin, 2016). The differences in findings may be due to the different species used during the experiment and the different ways in which PD was modelled. Therefore, further research would be needed to determine the exact effect of an increased phosphorylation of 4E-BP1, caused by mutant LRRK2, on the onset of PD. Furthermore, the studies have primarily modelled familial PD, so further research would be needed to determine the role of 4E-BP1 modification in sporadic cases of PD.
Moreover, Drosophila melanogaster is commonly used to model neurodegenerative diseases such as PD through the expression of alpha-synuclein in Drosophila models (Aryal and Lee, 2019). This allowed for the causal relationship between PD abnormalities such as dopaminergic cell degeneration, Lwey body formation, and locomotion dysfunction to be elucidated (Aryal and Lee, 2019). Furthermore, Drosophila and mammalian cell-cycle machinery and mitogenic signalling pathways, including the TOR pathway, are substantially conserved and TOR activity can be similarly measured in Drosophila by detecting phosphorylation of S6k at Thr398, the corresponding site in the Drosophila protein, which makes Drosophila a useful model in in vivo PD research (Khurana et al., 2006). However, in some less common LRRK2 mutations (R1441C, Y1699C, and I2020T), which frequently do not exhibit Lewy Body pathology, the Drosophila models become less accurate representations of the PD progression in such patients (Martin et al., 2014; Martin, 2016). Therefore, further research into investigating and modelling cases of PD that do not exhibit Lewy Body pathology would be useful in determining the role of LRRK2 mutations in different progressions of PD.
Notably, a different study reported that pathogenic LRRK2 represses the activity of let-7 and miR-184, which results in the upregulation of the transcription factors e2f1 and dp1 (Gehrke et al., 2010; Martin, 2016). This suggested that mutant LRRK2 has inhibitory effects on miRNA function (Gehrke et al., 2010; Martin, 2016). The inhibitory effects of LRRK2 on miRNA function are proposed through two mechanisms. The first hypothesis suggests that pathogenic LRRK2 leads to decreased levels of Drosophila Argonaute-1, a key component of the RNA-induced silencing complex, by an unknown mechanism (Gehrke et al., 2010; Martin, 2016). The second hypothesis suggests that mutant LRRK2 impairs the function of Argonaute-1 by associating it with phospho-4E-BP1 (Gehrke et al., 2010; Martin, 2016).
The LRRK2 mutation is associated with PD in patients. Three hypotheses about the relation between LRRK2 mutations and the development of PD have been suggested. Firstly, LRRK2 mutations were found to lead to neuronal enhanced kinase activity, that results in an increased phosphorylation of the ribosomal protein s15, and this stimulated both cap-dependent and cap-independent mRNA translation in an s15-dependent manner, which was associated with neuronal toxicity (Danilo, 2019; Martin, 2016). Secondly, the loss of normal LRRK2 leads to downregulated TOR signalling, drives apoptosis in dopaminergic neurons, and results in the neurodegenerative aspect of PD (Khurana et al., 2006; Martin, 2016). Thirdly, pathogenic LRRK2 represses the activity of let-7 and miR-184, which results in the upregulation of the transcription factors e2f1 and dp1 and consequently inhibits miRNA functions. It is important to note that there are limitations to the models used to investigate PD, such as the expression of alpha-synuclein in Drosophila models of PD being of limited use in modelling cases of PD that do not exhibit Lewy Body pathology. Therefore, further research that investigates the relationship between LRRK2 mutations and PD would be beneficial in bettering the current understanding of PD.
The PINK1 mutation
The PINK1 mutation is a mutation in the PTEN Induced Kinase 1 (PINK1) gene. It is associated with an increased risk of developing an early onset form of Parkinson’s disease, because this gene is required to make the protein PTEN induced putative kinase 1. This protein is required for the protection of mitochondria from damage and it also keeps the damage from replicating when cells reproduce. This process is known as mitophagy, whereby the mitochondria are selectively degraded by autophagy. (Malpartida et al., 2020). This lack of protection leads to damaged cells, then damaged neurons, and eventually causes Parkinson’s (Behring et al.). PINK1 mutations cause a decrease in its activity, which leads to an accumulation of mitochondrial damage and dysfunction. This ultimately leads to reduced energy production, an abnormal neuronal apoptosis (In a normal neuron, the apoptosis of cells does occur in a certain frame of time whereby the cell orchestrates its own death. Any apoptosis occurring after this frame of time is physiologically inappropriate and can lead to neurodegenerative disorders such as Parkinson’s) (Cavallaro), and the classic symptoms of Parkinson’s disease (Erekat et al., 2018).
Mutations in mitochondrial PINK1 are associated with recessive Parkinson’s disease. Notably, PINK1 causes apoptosis, abnormal mitochondrial morphology, impaired dopamine release and motor deficits. Therefore, it is hypothesized that motor symptoms of Parkinson’s are associated with this mutation. However, the fundamental mechanism underlying these phenotypes remains unknown as of now (Morais et al., 2009).
Although mutations or variants in many genes increase the susceptibility to Parkinson’s disease, only a handful of monogenic causes of parkinsonism have been identified. Biochemical and genetic studies reveal that the products of two genes that are mutated in autosomal recessive parkinsonism, PINK1 and Parkin, normally work together in the same pathway to govern mitochondrial quality control, bolstering previous evidence that mitochondrial damage is involved in Parkinson’s disease. PINK1 accumulates on the outer membrane of damaged mitochondria, activates Parkin’s E3 ubiquitin ligase activity, and recruits Parkin to the dysfunctional mitochondria. Then, Parkin ubiquitinates outer mitochondrial membrane proteins to trigger selective autophagy (Pickrell and Youle et al., 2015).
The PINK1 gene is associated with the onset of PD in two ways. Firstly, by mitochondrial dysfunction whereby the mitochondria in dopaminergic neurons become less efficient. This can lead to a decrease in dopamine production, which has been linked to the display of PD symptoms. Mitochondrial dysfunction is a hallmark of PD, and the PINK1 protein is thought to play an important role in mitigating this dysfunction. In normal cells, damaged mitochondria are targeted for destruction through mitophagy, which is regulated by the PINK1 protein. However, in cells with a mutated PINK1 gene, this process is impaired, leading to the accumulation of damaged and dysfunctional mitochondria.
The accumulation of dysfunctional mitochondria can lead to a range of cellular abnormalities, including increased production of reactive oxygen species (ROS), disruption of cellular energy metabolism, and the activation of cell death pathways. These abnormalities can contribute to the loss of dopaminergic neurons in the substantia nigra, as these cells are particularly sensitive to mitochondrial dysfunction and oxidative stress. The reasons for this however are still unknown.
In addition to its role in mitophagy, the PINK1 protein is also thought to play a role in the regulation of protein aggregation, another hallmark of PD. Specifically, the PINK1 protein has been shown to inhibit the aggregation of alpha-synuclein, a protein that is found in Lewy bodies, the characteristic protein aggregates that are a hallmark of PD. In cells with a mutated PINK1 gene, this inhibition is impaired. This is because the PINK1 gene has a link to the gene parkin which has a role in the clearance of insoluble protein aggregates via macroautophagy and has also been implicated by more recent studies. Paradoxically, like alpha-synuclein, parkin is also prone to misfolding, especially in the presence of age-related stress. Similarly, protein misfolding can also affect the function of other key PD-linked genes such as DJ-1, PINK1, and perhaps also LRRK2. This similarly causes the dysfunction of the PINK1 gene (Tan et al., 2009).
RNAi: A Promising Treatment to Parkinson’s Disease
A currently studied treatment for Parkinson’s disease is RNAi which stands for RNA interference. RNAi refers to a biological process where small pieces of RNA prevent protein translation by degrading mRNA. MRNA, which stands for messenger RNA, is a single-stranded RNA with the genetic information needed to make proteins (National Human Genome Research Institute, 2020). In protein synthesis, mRNA is exported from the nucleus into the cytoplasm, where ribosomes read the genetic code of the mRNA to form polypeptide chains that fold into proteins. However, RNAi inhibits the mRNA before it is translated into the ribosome (Xu, Jiang and Huang, 2019).
The small pieces of RNA that enable RNA interference are siRNA or microRNA, derived from dsRNA (double-stranded RNA). DsRNA don’t naturally exist in the cell; they get into the cell through multiple ways, such as viral infections and transfection, which is when researchers introduce dsRNA molecules into cells in the laboratory (Matsumoto and Seya, 2013). The process of RNAi starts when the double-stranded precursors of siRNAs or microRNAs bind to Dicer, a protein that cuts the RNA into short segments, approximately 21 nucleotides long. These short segments then bind to argonaut protein, where one strand of the RNA is selected. The combination of the RNA, argonaut, and other proteins is called the RNA-induced silencing complex (RISC). SiRNA or microRNA directs RISC to the targeted mRNA. Once bound, RISC can cleave the mRNA molecule or block its translation into protein by preventing ribosomes from accessing the mRNA. In the case of siRNA, the targeting is precise because it is determined by base pairing between siRNA and the target mRNA, which has perfect complementarity. In the case of microRNA, the pairing is not as precise because only a part of the microRNA, called the seed, pairs with the targeted mRNA. This type of pairing makes the degrading process much bigger by targeting a larger number of similar sequences (UMass Chan Medical School, 2013).
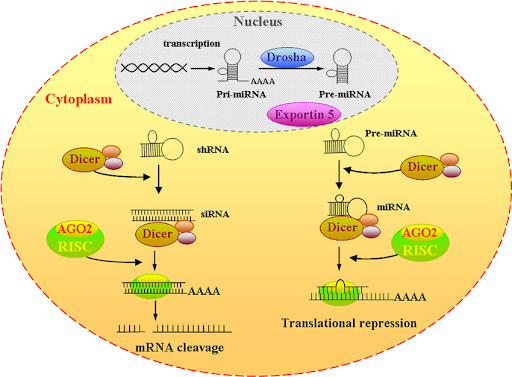
Figure 2 – Diagram showing the simplified pathways of siRNA and microRNA to inhibit mRNA. [Taken from (Tan, Yang and Ji, 2009)]
RNAi therapeutics is a class of drugs that utilises specially designed siRNA or microRNA to target and silence specific genes that cause or contribute to disease. RNAi can also target genes that improve the levels of other non-disease-causing proteins involved in the disease pathway. Also, when a protein’s function is still unknown, silencing genes help scientists observe what happens in its absence, and from there, learn what it does. Using RNAi technology, scientists have identified genes that represent new therapeutic targets for treating Parkinson’s disease (Bumcrot et al., 2006).
In order to target and disable mRNA-involved pathways implicated in PD, siRNAs must be specifically and efficiently delivered into the cell. RNAi therapeutics have different approaches to enabling the delivery of siRNA to the cell. One secure approach is lipid nanoparticles, which are chemically synthesised multicomponent lipid formulations. Lipid nanoparticles encapsulate siRNAs and protect them from degradation by ubiquitous nucleases until they enter the cell’s cytoplasm (Hou et al., 2021).
Problems with mitochondria play a significant role in the pathogenesis of PD. Some cases of PD have been linked to mutations in the gene that codes for parkin, a protein that tags damaged mitochondria as waste. Mutations in parkin prevent tagging, accumulating unhealthy mitochondria in the body, affecting cellular pathways, and leading to cellular damage and death (Moon and Paek, 2015). Scientists at the NIH used RNAi to identify genes that help parkin tag damaged mitochondria. The researchers found that turning off some genes, including BAG4 and SIAH3, enhanced tagging. Further experiments supported that these genes may be new targets for treating Parkinson’s and other neurological disorders (National Institutes of Health, 2016)
These results appear promising, as enhancing tagging in parkin can be a way to treat mitochondrial accumulation in PD. A possible treatment could be designing siRNA for BAG4 and SIAH3 that would travel to the targeted cell using lipid nanoparticles. Then the siRNA would degrade the targeted mRNA for BAG4 and SIAH3, which, in turn, enhances the tagging of parkin, causing a reduction of mitochondrial accumulation. However, the challenges of this study are that a treatment silencing these genes will only be effective for some patients since this mutation only occurs in some cases of PD. Additionally, there could be other mutations affecting mitochondrial accumulation, which would require different RNAi sequences, making this not a feasible solution.
RNAi research has also been done on SNCA, the gene responsible for making ɑ-synuclein. A study done on 8-month-old rats focused on silencing SNCA through the use of an adeno-associated virus (AAV), which is a non-enveloped virus that can be engineered to deliver DNA to target cells (Naso et al., 2017). The rats were injected with either an RNAi vector targeting the SNCA mRNA transcript (AAV-sh(SNCA)) or a control vector (AAV-sh(Ctrl)) into the substantia nigra of the animal. After 12 months, SNCA mRNA expression in the substantia nigra of the rats that received (AAV-sh(SNCA)) decreased by approximately 90%, and α-synuclein immunoreactivity decreased by more than 70% relative to the control side. There was no evidence of neurodegeneration in the substantia nigra 12 months after injection with either vector. The results conclude long-term a-synuclein knockdown in the substantia nigra with no side effects, which is encouraging for targeting alpha-synuclein in patients with Parkinson’s disease (Zharikov et al., 2019).
It is important to mention that RNAi targeting the SNCA gene is limited to the area of movement in PD. Restoring levels of dopamine in the substantia nigra could have beneficial effects on the motor symptoms of the disease. However, other treatments would be needed to target the non-motor and psychiatric symptoms that result from different areas of the brain that are not related to movement. RNAi-based therapies for Parkinson’s disease are still in the early stages of development and face several challenges, such as off-target effects and stimulation of the immune response. Clinical trials will be needed to determine the safety and efficacy of RNAi-based therapies for Parkinson’s disease in humans. However, it has a lot of potential as it continues to be explored and developed in Parkinson’s and numerous other diseases (Prabhune, 2019).
CRISPR
What is CRISPR?
CRISPR-Cas9 are naturally existing proteins mainly found in bacterial immune systems that can be sourced from Streptococcus pyogenes. Researchers can utilize single guide RNA (sgRNA), derived from a linkage between crRNA and tracrRNA, to isolate the specific sequence of DNA that requires editing. Guidance RNA that has been synthesised in a complementary sequence to the DNA of interest will bind to the section of the DNA that Cas9 will cut out. Cas9 has two domains with each domain responsible for cutting a strand of DNA. After the DNA is severed, it can undergo two forms of repair: homology-directed repair (HDR), or non-homologous end joining (NHEJ) in which the nucleotide combination can be manually changed. HDR naturally occurs after a double strand break, which the CRISPR/Cas9 system creates, where DNA damage is repaired by pasting a sequence provided by a donor template. In NHEJ, DNA repair after CRISPR’s edits occurs without a homologous template in which the break ends are directly ligated (Li et al., 2017).
Past Tests for CRISPR/Cas9
Past attempts at using CRISPR as a cure for diseases have proven to be successful, which shows possible CRISPR applications to PD. For instance, used CRISPR/Cas9 to fix a gene defect in male and female human zygotes that were carriers for hypertrophic cardiomyopathy. The end result of this procedure was a sustainable human embryo that was free of the disease. Thus, providing hope that gene therapy for germline mutations is feasible and a possible treatment for hereditary PD (Ma et al., 2017).
Possible Applications for Parkinson’s
CRISPR can be used to edit out aforementioned germline genes that are known to cause hereditary Parkinson’s. For instance, the aforementioned LRRK2 germline mutation is currently being targeted with a gRNA that contains nucleotides homologous to the G6055 site. The Cas9 protein could sever the DNA strand at the LRRK2 mutation at the G6055 location, and HDR will occur immediately after replacing LRRK2 with a segment of non-PD correlated gene (Vermilyea et al., 2020). As research in PD inducing mutations and CRISPR advances to clinical use, it has potential implications in preventing hereditary PD. Although this may sound revolutionary, some germline mutations are not proven to cause PD but rather only show a correlation to PD. Therefore, this method may not be guaranteed to prevent PD. In addition, according to Klein and Westenberger (2012), hereditary PD only consists of 15% of all PD patients, thus investing in this may risk large spending for only treating a fraction of PD patients.
Another strategy for utilizing CRISPR to prevent Parkinson’s disease is altering a segment of DNA in neurons responsible for dopamine secretion. Researchers may use CRISPR to change existing neurons to create more dopamine, or replace diseased neurons with healthy ones.
It is also possible to use CRISPR to transfer dopamine instructions to STEM cells where dopamine could be harvested and given to people with Parkinson’s Disease to decrease symptom severity (Yang et al., 2019). The early stages of research for this treatment has already begun as researchers started transplanting dopamine-producing neurons made from stem cells using CRISPR animals with Parkinson’s disease. Results show that the animals’ motor symptoms significantly improve, which correlated with a considerable increase in dopamine.
Personalizing PD treatment with iPSCs and Single cell RNA sequencing:
Induced pluripotent stem cells (iPSC) are being utilized in many techniques, because this personalizes the treatment, and consequently reduces the risk of an immune rejection. Single cell sequencing is a technique that individualizes research to comprehend exactly how PD has affected each patient and how to treat it. The first study on this sequencing was issued in 2009, and since then researchers all over the world have been able to make many new discoveries thanks to this research method. It consists of detecting and quantitatively analysing RNA on a genetic level in a given specimen (Haque et al., 2017). It is also important to note that single cell sequencing is much more reliable than bulk RNA sequencing since each cell’s quantitative expression is calculated, rather than just the average expression (Kim and Daadi, 2019). It can be used with in vivo and in vitro models, which allow researchers to track, observe and investigate the progression of the study, one cell at a time. To get stem cells, the researchers extract the diseased cell they need from each PD patient they are treating. Then, they can proceed to transform it, and reinject it into a living model, like a mouse. This allows scientists to explore how the cell reacts to different stimuli or genetic mutations. The knowledge gained from this method will help further understand the mutations, and their effects on surrounding cells (Kim and Daadi, 2019).
Single cell sequencing can be used to determine how to modify induced pluripotent stem cells (iPSC), which are obtained from blood or skin cells. Once given back to the patient, the corrected cells can compensate for the damaged ones. The blood and skin cells can be conditioned to return to an embryonic-like pluripotent state. This allows it to have the ability to become any type of human cell, including neurons, and many more. They can then be used for therapeutic purposes. The use of iPSC is more patient specific, they can be programmed to accommodate specific genetic risk factors to assess genetic sub-populations. At present, it is the most promising cell therapy for slowing or stopping PD progression (Stoddard-Bennett and Reijo Pera, 2019). Since iPSCs can become neurons, they could be used for gene therapy in PD, and can be used to study and learn more about the mutated genes. Researchers can learn how the genes function and how the surrounding genes react to these mutations. Knowing how the disease starts is the first step to knowing how to cure or counter it. Single cell sequencing is not a cure in itself, but accompanied by other techniques like using iPSC, there are so many opportunities to find cures for PD. Single cell RNA sequencing research is now worldwide, so many different cells are being tested to evaluate and study their effects on the development of PD (Novak et al., 2022).
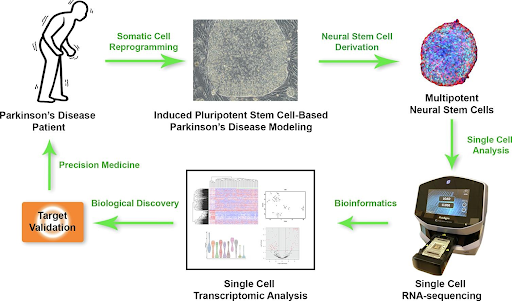
(Kim and Daadi, 2019)
This graphic shows a possible path from diagnosis to cure when using single-cell RNA sequencing and iPSC modulation. The first step is extracting a sample of the cells affected by mutated genes (Kim and Daadi, 2019). Once the stem cell has been modelled and studied, the next step is Neural Stem Cell Derivation which consists of covering mouse embryonic fibroblasts (MEF) with blastocysts which are aggregations of stem cells very early in the pregnancy, followed by its growth and progression in the embryo (Stojkovic et al., 2004). Multipotent stem cells have been partially differentiated, but they can still become a wide variety of cells. They have many great uses, and they are the main contributors in helping with tissue repair, protection, and development (Sobhani et al., 2017). After analyzing the single cells, the researchers can proceed with the sequencing, from which they will get bioinformatics, which is information on each of the cells individually. This is a lot more accurate and helpful, because each cell is studied on its own and has its own expression and information. Approaching the end of this process, they start single cell transcriptomic analysis, which is to extract the most important transcription factors from a cell. Followed by target modulation, which is just a verification that the cell, and its modulation, are actually involved with and will help cure or treat the disease (Haque et al., 2017). Finally, the patient can be treated with any of the procedures discussed earlier in the paper, depending on what option is best for the patient.
One of the most promising trials is the JMA-IIA00384, UMIN000033564, where they are using iPSC-derived dopaminergic progenitors (DAPs) to treat the disease. The importance of the iPSCs is that they act as DaNs which PD patients lack. This trial has a much higher risk for the patient, a non-human primate, since DAPs are being replaced rather than supplying neurotrophic support or immunomodulation. This technique was proven safe and effective in non-human primates, which is very promising. The grafted cells from this trial seem to remain alive for a very long time, perhaps until the death of the patient. In this trial, they would differentiate the cells into DAPs and then transplant them. This trial started the final phase, which is clinical trials on humans (Doi et al., 2020). It was the first iPSC human transplant and represented an immense scientific advancement.
The main point to keep in mind is that this treatment, cell replacement therapy, helps the symptoms, but does not slow the disease’s progression. Contrarily to a disease modifying therapy, which only stops the disease pathology, but not the symptoms. In order to fully treat and heal this neurodegenerative disease, the two techniques need to be combined (Takahashi, 2020).
Conclusion
RNAi, CRISPR, and single-cell sequencing are technologies that demonstrate potential for PD treatment. RNAi has shown promising results in silencing gene expression associated with PD. However, it is important to note that further research is needed to determine the safety and effectiveness of using RNAi treatments in human patients with PD. Furthermore, the contemporary development of CRISPR for germline disease prevention could be of possible future use in preventing hereditary PD and the treatment of PD. Moreover, single-cell sequencing is a technique that can be used to individualize the other treatments and diagnosis methods. Studying iPSCs, and a treatment involving the modulation of their mutated versions before giving them back to the patient through a gene therapy such as CRISPR or RNAi can aid the treatment of PD. These techniques hold great potential for developing more precise and effective PD treatments and could serve as a baseline for developing treatments for other neurodegenerative disorders. Although further research is required to optimize the use of gene therapies in treating PD, RNAi, CRISPR, and single-cell sequencing technologies hold the potential to improve current PD treatments.
Works Cited
Aryal, B., & Lee, Y. (2019). Disease model organism for parkinson disease: <i>Drosophila melanogaster</i>. BMB Reports, 52(4), 250-258. https://doi.org/10.5483/BMBRep.2019.52.4.204
Behring, S., et al. “How Mutations in the PINK1 Gene May Lead to Parkinson’s Disease.” Healthline, 23 June 2022, www.healthline.com/health/parkinsons/pink1-gene-parkinsons Accessed 23 Feb. 2023.
Blauwendraat, C., Reed, X., Krohn, L., Heilbron, K., Bandres-ciga, S., Tan, M., Gibbs, J. R., Hernandez, D. G., Kumaran, R., Langston, R., Bonet-ponce, L., Alcalay, R. N., Hassin-baer, S., Greenbaum, L., Iwaki, H., Leonard, H. L., Grenn, F. P., Ruskey, J. A., Sabir, M., . . . Siitonen, A. (2019). Genetic modifiers of risk and age at onset in GBA associated parkinson’s disease and lewy body dementia. Brain, 143(1), 234-248. https://doi.org/10.1093/brain/awz350
Bosgraaf, L., & Van haastert, P. J.m. (2003). Roc, a ras/gtpase domain in complex proteins. Biochimica Et Biophysica Acta (BBA) – Molecular Cell Research, 1643(1-3), 5-10. https://doi.org/10.1016/j.bbamcr.2003.08.008
Bouhouche, A., Tibar, H., Ben el haj, R., El bayad, K., Razine, R., Tazrout, S., Skalli, A., Bouslam, N., Elouardi, L., Benomar, A., Yahyaoui, M., & Regragui, W. (2017). <i>LRRK2</i> g2019s mutation: Prevalence and clinical features in moroccans with parkinson’s disease. Parkinson’s Disease, 2017, 1-7. https://doi.org/10.1155/2017/2412486
Bumcrot, D., Manoharan, M., Koteliansky, V. and Sah, D.W.Y. (2006). RNAi therapeutics: a potential new class of pharmaceutical drugs. Nature Chemical Biology, [online] 2(12), pp.711–719. doi:https://doi.org/10.1038/nchembio839
Cavallaro, S. “Cracking the Code of Neuronal Apoptosis and Survival.” Cell Death & Disease, vol. 6, no. 11, Nov. 2015, pp. e1963–e1963, https://doi.org/10.1038/cddis.2015.309.
Correddu, D., & Leung, I. K.h. (2019). Targeting mRNA translation in parkinson’s disease. Drug Discovery Today, 24(6), 1295-1303. https://doi.org/10.1016/j.drudis.2019.04.003
Deng, J., Lewis, P. A., Greggio, E., Sluch, E., Beilina, A., & Cookson, M. R. (2008). Structure of the ROC domain from the parkinson’s disease-associated leucine-rich repeat kinase 2 reveals a dimeric gtpase. Proceedings of the National Academy of Sciences, 105(5), 1499-1504. https://doi.org/10.1073/pnas.0709098105
Gehrke, S., Imai, Y., Sokol, N., & Lu, B. (2010). Pathogenic lrrk2 negatively regulates microRNA-mediated translational repression. Nature, 466(7306), 637-641. https://doi.org/10.1038/nature09191
Harraz, M. M., Dawson, T. M., & Dawson, V. L. (2011). MicroRNAs in parkinson’s disease. Journal of Chemical Neuroanatomy, 42(2), 127-130. https://doi.org/10.1016/j.jchemneu.2011.01.005
Healy, D. G., Falchi, M., O’sullivan, S. S., Bonifati, V., Durr, A., Bressman, S., Brice, A., Aasly, J., Zabetian, C. P., Goldwurm, S., Ferreira, J. J., Tolosa, E., Kay, D. M., Klein, C., Williams, D. R., Marras, C., Lang, A. E., Wszolek, Z. K., Berciano, J., . . . Wood, N. W. (2008). Phenotype, genotype, and worldwide genetic penetrance of lrrk2-associated parkinson’s disease: A case-control study. The Lancet Neurology, 7(7), 583-590. https://doi.org/10.1016/S1474-4422(08)70117-0
Howells, D.w., Porritt, M.j., Wong, J., Batchelor, P.e., Kalnins, R., Hughes, A.j., & Donnan, G.a. (2000). Reduced BDNF mRNA expression in the parkinson’s disease substantia nigra. Experimental Neurology, 166(1), 127-135. https://doi.org/10.1006/exnr.2000.7483
Hou, X., Zaks, T., Langer, R. and Dong, Y. (2021). Lipid nanoparticles for mRNA delivery. Nature Reviews Materials, 6. doi:https://doi.org/10.1038/s41578-021-00358-0
Johns Hopkins Medicine. “Targeting Parkinson’s-linked protein could neutralize two of the disease’s causes: Mouse and human cell studies confirm a common link between proteins and loss of dopamine-making brain cells.” ScienceDaily. ScienceDaily, 1 February 2017. <www.sciencedaily.com/releases/2017/02/170201110632.htm>.
Khurana, V., Lu, Y., Steinhilb, M. L., Oldham, S., Shulman, J. M., & Feany, M. B. (2006). TOR-Mediated cell-cycle activation causes neurodegeneration in a drosophila tauopathy model. Current Biology, 16(3), 230-241. https://doi.org/10.1016/j.cub.2005.12.042
Klein, C. and Westenberger, A. (2012). Genetics of Parkinson’s Disease. Cold Spring Harbor Perspectives in Medicine, [online] 2(1), pp.a008888–a008888. doi:https://doi.org/10.1101/cshperspect.a008888.
Knipe, H., & Gaillard, F. (2011). Parkinson disease. In Radiopaedia.org. Radiopaedia.org. https://doi.org/10.53347/rID-13027
Kumar, A., Greggio, E., Beilina, A., Kaganovich, A., Chan, D., Taymans, J.-M., Wolozin, B., & Cookson, M. R. (2010). The parkinson’s disease associated lrrk2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation. PLoS ONE, 5(1), e8730. https://doi.org/10.1371/journal.pone.0008730
Li, X.-J., Tu, Z., Yang, W. and Li, S. (2017). CRISPR: Established Editor of Human Embryos? Cell stem cell, [online] 21(3), pp.295–296. doi:https://doi.org/10.1016/j.stem.2017.08.007.
Lopez, G., Lazzeri, G., Rappa, A., Isimbaldi, G., Cribiù, F. M., Guerini-rocco, E., Ferrero, S., Vaira, V., & Di fonzo, A. (2020). Comprehensive genomic analysis reveals the prognostic role of lrrk2 copy-number variations in human malignancies. Genes, 11(8), 846. https://doi/org/10.3390/genes11080846
Ma, H., Marti-Gutierrez, N., Park, S.-W., Wu, J., Lee, Y., Suzuki, K., Koski, A., Ji, D., Hayama, T., Ahmed, R., Darby, H., Van Dyken, C., Li, Y., Kang, E., Park, A.-R., Kim, D., Kim, S.-T., Gong, J., Gu, Y. and Xu, X. (2017). Correction of a pathogenic gene mutation in human embryos. Nature, [online] 548(7668), pp.413–419. doi:https://doi.org/10.1038/nature23305.
Martin, I. (2016). Decoding parkinson’s disease pathogenesis: The role of deregulated mRNA translation. Journal of Parkinson’s Disease, 6(1), 17-27. https://doi.org/10.3233/JPD-150738
Martin, I., Kim, J. W., Dawson, V. L., & Dawson, T. M. (2014). LRRK2 pathobiology in parkinson’s disease. Journal of Neurochemistry, 131(5), 554-565. https://doi.org/10.1111/jnc.12949
Matsumoto, M. and Seya, T. (2013). Double-Stranded RNA – an overview | ScienceDirect Topics. [online] www.sciencedirect.com. Available at: https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/double-stranded-rna#:~:text=Double%2Dstranded%20RNA%20is%20generated [Accessed 25 Feb. 2023].
Moon, H.E. and Paek, S.H. (2015). Mitochondrial Dysfunction in Parkinson’s Disease. Experimental Neurobiology, [online] 24(2), p.103. doi:https://doi.org/10.5607/en.2015.24.2.103.
Morais, Vanessa A, et al. “Parkinson’s Disease Mutations in PINK1 Result in Decreased Complex I Activity and Deficient Synaptic Function.” EMBO Molecular Medicine, vol. 1, no. 2, 1 May 2009, pp. 99–111, www.ncbi.nlm.nih.gov/pmc/articles/PMC3378121/, https://doi.org/10.1002/emmm.200900006. Accessed 19 Nov. 2020.
Morais VA, Verstreken P, Roethig A, Smet J, Snellinx A, Vanbrabant M, Haddad D, Frezza C, Mandemakers W, Vogt-Weisenhorn D, Van Coster R, Wurst W, Scorrano L, De Strooper B. Parkinson’s disease mutations in PINK1 result in decreased Complex I activity and deficient synaptic function. EMBO Mol Med. 2009 May;1(2):99-111. doi: 10.1002/emmm.200900006. PMID: 20049710; PMCID: PMC3378121.
Naso, M.F., Tomkowicz, B., Perry, W.L. and Strohl, W.R. (2017). Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs, 31(4), pp.317–334. doi:https://doi.org/10.1007/s40259-017-0234-5.
National Human Genome Research Institute (2020). Messenger RNA (mRNA). [online] Genome.gov. Available at: https://www.genome.gov/genetics-glossary/messenger-rna.
National Institutes of Health (NIH). (2016). Gene-silencing study finds new targets for Parkinson’s disease. [online] Available at: https://www.nih.gov/news-events/news-releases/gene-silencing-study-finds-new-targets-parkinsons-disease.
Norat, Pedro, et al. “Mitochondrial Dysfunction in Neurological Disorders: Exploring Mitochondrial Transplantation.” Npj Regenerative Medicine, vol. 5, no. 1, 23 Nov. 2020, https://doi.org/10.1038/s41536-020-00107-x.
parkinsonsnewstoday.com/news/research-reveals-how-park7-gene-mutations-affecting-dj-1-protein-cause-early-onset-parkinsons/#:~:text=The%20PARK7%20gene%20encodes%20the%20instructions%20for%20DJ-1%2C. Accessed 23 Feb. 2023.
Parkinson disease. (2012, May 1). MedlinePlus. Retrieved February 9, 2023, from https://medlineplus.gov/genetics/condition/parkinson-disease/#causes
“Parkin (Protein).” Wikipedia, 19 Oct. 2022, en.wikipedia.org/wiki/Parkin_(protein). Accessed 23 Feb. 2023.
Perni, Michele, et al. “Comparative Studies in the A30P and A53T α-Synuclein C. Elegans Strains to Investigate the Molecular Origins of Parkinson’s Disease.” Frontiers in Cell and Developmental Biology, vol. 9, 22 Mar. 2021, https://doi.org/10.3389/fcell.2021.552549. Accessed 22 Mar. 2021.
PhD, Steve Bryson. “How PARK7 Gene Mutations Cause Early Onset Parkinson’s Revealed.” Parkinson’s News Today, 15 Sept. 2020,
Pickrell, Alicia M., and Richard J. Youle. “The Roles of PINK1, Parkin, and Mitochondrial Fidelity in Parkinson’s Disease.” Neuron, vol. 85, no. 2, Jan. 2015, pp. 257–273,
Pozdyshev, D. V., Zharikova, A. A., Medvedeva, M. V., & Muronetz, V. I. (2021). Differential analysis of a-to-i mRNA edited sites in parkinson’s disease. Genes, 13(1), 14. https://doi.org/10.3390/genes13010014
Prabhune, M. (2019). Synthego | Full Stack Genome Engineering. [online] Synthego.com. Available at: https://www.synthego.com/blog/rnai-vs-crispr-guide.
Salat, D., & Tolosa, E. (2013). Levodopa in the treatment of parkinson’s disease: Current status and new developments. Journal of Parkinson’s Disease, 3(3), 255-269. https://doi.org/10.3233/JPD-130186
Serganov, A. (2003). Ribosomal protein s15 represses its own translation via adaptation of an rRNA-like fold within its mRNA. The EMBO Journal, 22(8), 1898-1908. https://doi.org/10.1093/emboj/cdg170
Tain, L. S., Mortiboys, H., Tao, R. N., Ziviani, E., Bandmann, O., & Whitworth, A. J. (2009). Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nature Neuroscience, 12(9), 1129-1135. https://doi.org/10.1038/nn.2372
Tan, P.-H., Yang, L.-C. and Ji, R.-R. (2009). Therapeutic Potential of RNA Interference in Pain Medicine. The Open Pain Journal, 2(1), pp.57–63. doi:https://doi.org/10.2174/1876386300902010057.
“The Genetic Link to Parkinson’s Disease.” Www.hopkinsmedicine.org, 10 Apr. 2022, www.hopkinsmedicine.org/health/conditions-and-diseases/parkinsons-disease/the-genetic-link-to-parkinsons-disease#:~:text=Targeting%20Parkinson%E2%80%99s-Linked%20Protein%20Could%20Neutralize%202%20of%20the. Accessed 18 Feb. 2023.
“The Genetic Link to Parkinson’s Disease.” Www.hopkinsmedicine.org, The Johns Hopkins University, The Johns Hopkins Hospital, and Johns Hopkins Health System., www.hopkinsmedicine.org/health/conditions-and-diseases/parkinsons-disease/the-genetic-link-to-parkinsons-disease.
UMass Chan Medical School. (2013). What is RNAi – RNAi Biology | UMass Medical School. [online] Available at: https://www.umassmed.edu/rti/biology/rna/how-rnai-works/.
Vermilyea, S.C., Babinski, A., Tran, N., To, S., Guthrie, S., Kluss, J.H., Schmidt, J.K., Wiepz, G.J., Meyer, M.G., Murphy, M.E., Cookson, M.R., Emborg, M.E. and Golos, T.G. (2020). In Vitro CRISPR/Cas9-Directed Gene Editing to Model LRRK2 G2019S Parkinson’s Disease in Common Marmosets. Scientific Reports, [online] 10(1), p.3447. doi:https://doi.org/10.1038/s41598-020-60273-2.
Wikipedia Contributors. “Mitophagy.” Wikipedia, Wikimedia Foundation, 10 Apr. 2019, en.wikipedia.org/wiki/Mitophagy. Accessed 24 Apr. 2019.
www.hopkinsmedicine.org. (n.d.). The Genetic Link to Parkinson’s Disease. [online] https://www.hopkinsmedicine.org/health/conditions-and-diseases/parkinsons-disease/the-genetic-link-to-parkinsons-disease.
www.ncbi.nlm.nih.gov/pmc/articles/PMC4764997/, https://doi.org/10.1016/j.neuron.2014.12.007.
www.sciencedirect.com. (n.d.). Double Stranded RNA – an overview | ScienceDirect Topics. [online] Available at: https://www.sciencedirect.com/topics/pharmacology-toxicology-and-pharmaceutical-science/double-stranded-rna [Accessed 24 Feb. 2023].
Xu, W., Jiang, X. and Huang, L. (2019). RNA Interference Technology. Comprehensive Biotechnology, [online] pp.560–575. doi:https://doi.org/10.1016/B978-0-444-64046-8.00282-2.
Yang, H., Hao, D., Liu, C., Huang, D., Chen, B., Fan, H., Liu, C., Zhang, L., Zhang, Q., An, J. and Zhao, J. (2019). Generation of functional dopaminergic neurons from human spermatogonial stem cells to rescue parkinsonian phenotypes. Stem Cell Research & Therapy, 10(1). doi:https://doi.org/10.1186/s13287-019-1294-x.
Zharikov, A., Bai, Q., De Miranda, B.R., Van Laar, A., Greenamyre, J.T. and Burton, E.A. (2019). Long-term RNAi knockdown of α-synuclein in the adult rat substantia nigra without neurodegeneration. Neurobiology of Disease, 125, pp.146–153. doi:https://doi.org/10.1016/j.nbd.2019.01.004.
Doi, D., Magotani, H., Kikuchi, T., Ikeda, M., Hiramatsu, S., Yoshida, K., Amano, N., Nomura, M., Umekage, M., Morizane, A. and Takahashi, J. (2020). Pre-clinical study of induced pluripotent stem cell-derived dopaminergic progenitor cells for Parkinson’s disease. Nature Communications, [online] 11(1), p.3369. doi:https://doi.org/10.1038/s41467-020-17165-w.
Haque, A., Engel, J., Teichmann, S.A. and Lönnberg, T. (2017). A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Medicine, 9(1). doi:https://doi.org/10.1186/s13073-017-0467-4.
Kim, J. and Daadi, M.M. (2019). Non-cell autonomous mechanism of Parkinson’s disease pathology caused by G2019S LRRK2 mutation in Ashkenazi Jewish patient: Single cell analysis. Brain Research, 1722, p.146342. doi:https://doi.org/10.1016/j.brainres.2019.146342.
Novak, G., Kyriakis, D., Grzyb, K., Bernini, M., Rodius, S., Dittmar, G., Finkbeiner, S. and Skupin, A. (2022). Single-cell transcriptomics of human iPSC differentiation dynamics reveal a core molecular network of Parkinson’s disease. Communications Biology, 5(1). doi:https://doi.org/10.1038/s42003-021-02973-7.
Sobhani, A., Khanlarkhani, N., Baazm, M., Mohammadzadeh, F., Najafi, A., Mehdinejadiani, S. and Sargolzaei Aval, F. (2017). Multipotent Stem Cell and Current Application. Acta Medica Iranica, [online] 55(1), pp.6–23. Available at: https://pubmed.ncbi.nlm.nih.gov/28188938/#:~:text=Multipotent%20stem%20cells%20have%20the.
Stojkovic, M., Lako, M., Strachan, T. and Murdoch, A. (2004). Derivation, growth and applications of human embryonic stem cells. Reproduction, 128(3), pp.259–267. doi:https://doi.org/10.1530/rep.1.00243.
Stoddard-Bennett, T. and Reijo Pera, R. (2019). Treatment of Parkinson’s Disease through Personalized Medicine and Induced Pluripotent Stem Cells. Cells, [online] 8(1), p.26. doi:https://doi.org/10.3390/cells8010026.
Takahashi, J. (2020). iPS cell-based therapy for Parkinson’s disease: A Kyoto trial. Regenerative Therapy, [online] 13. doi:https://doi.org/10.1016/j.reth.2020.06.002.
Thaler, A., Ash, E., Gan-Or, Z., Orr-Urtreger, A. and Giladi, N. (2009). The LRRK2 G2019S Mutation as the Cause of Parkinson’s Disease in Ashkenazi Jews. Journal of Neural Transmission, 116(11), pp.1473–1482. doi:https://doi.org/10.1007/s00702-009-0303-0.